Datasets¶
Working with large-scale imaging data can be both challenging and messy, so Slideflow provides the Dataset class to assist with managing, splitting, filtering, and transforming your data for easy downstream use. Dataset organizes a set of image tiles extracted at a specific size, along with their associated slides and clinical annotations. Datasets are used for many Slideflow functions, and can quickly generate torch.utils.data.DataLoader and tf.data.Datasets objects that provide preprocessed slide images for external applications.
Dataset Sources¶
Datasets are comprised of one or more sources, which are a set of slides, Regions of Interest (if available), and any tiles extracted from these slides. You might choose to organize your data into separate sources if slides are organized into distinct locations on disk - for example, if you are using multiple sets of slides from different institutions, with data from each institution stored separately.
Loading a Dataset¶
Datasets can be created either from a Project - using the project’s dataset configuration file - or directly by providing paths to slides, annotations, and image tile destinations. In the next sections, we’ll take a look at how to create a Dataset with each method.
From a project¶
If you are working in the context of a Project, a dataset can be quickly created using Project.dataset(). A dataset can be loaded from a given Project with the following parameters:
tile_pxis the tile size, in pixelstile_umis the tile size, in microns (int) or magnification ('40x')sourcesis an optional list of dataset sources to use
import slideflow as sf
P = sf.load_project('/project/path')
dataset = P.dataset(tile_px=299, tile_um='10x', sources=['Source1'])
If sources is not provided, all available sources will be used.
Alternatively, you can accomplish the same by creating a Dataset object directly, passing in the project dataset configuration file to the config argument, and a path to the annotations file to annotations:
dataset = sf.Dataset(
config='config.json',
sources=['Source1'],
annotations='annotations.csv',
tile_px=299,
tile_um='10x'
)
Manually from paths¶
You can also create a dataset by manually supplying paths to slides, destination for image tiles, and clinical annotations. A single dataset source will be created from the provided arguments, which include:
tile_pxis the tile size, in pixelstile_umis the size in microns or magnificationslidesis the directory containing whole-slide imagesroiis the directory containing Regions of Interest *.csv filestfrecordsis the path to where image tiles should be stored in TFRecordstilesis the path to where image tiles should be stored as *.jpg imagesannotationsis either an annotations file (CSV) or Pandas DataFrame.
For example, to create a dataset from a set of slides, with a configured TFRecord directory and annotations provided via Pandas DataFrame:
import pandas as pd
# Create some clinical annotations
df = pd.DataFrame(...)
# Create a dataset
dataset = sf.Dataset(
slides='/slides',
tfrecords='/tfrecords',
annotations=df,
tile_px=299,
tile_um='10x'
)
When creating a Dataset manually from paths, tfrecords should be organized into subdirectories named according to tile size. Using the above example, the tfrecords directory should look like:
/tfrecords
└── 299px_10x
├── slide1.tfrecords
├── slide2.tfrecords
├── slide3.tfrecords
└── ...
Filtering¶
Datasets can be filtered through several mechanisms:
filters: A dictionary, where keys are clinical annotation headers and values are the variable states which should be included. All remaining slides are removed from the dataset.
filter_blank: A list of headers; any slide with a blank value in the clinical annotations in one of these columns will be excluded.
min_tiles: An
int; any tfrecords with fewer than this number of tiles will be excluded.
Filters can be provided at the time of Dataset creation by passing to the initializer:
dataset = Dataset(..., filters={'HPV_status': ['negative', 'positive']})
or by using the Dataset.filter() method:
dataset = dataset.filter(min_tiles=50)
Dataset Manipulation¶
A number of functions can be applied to Datasets to manipulate patient filters (Dataset.filter(), Dataset.remove_filter(), Dataset.clear_filters()), clip tfrecords to a maximum number of tiles (Dataset.clip()), or prepare mini-batch balancing (Dataset.balance()). The full documentation for these functions is given in the API. Each of these manipulations return an altered copy of the dataset for easy chaining:
dataset = dataset.balance('HPV_status').clip(50)
Each of these manipulations is performed in memory and will not affect data stored on disk.
Dataset Inspection¶
The fastest way to inspect a Dataset and the dataset sources loaded, number of slides found, clinical annotation columns available, and number of tiles extracted into TFRecords is the Dataset.summary() method.
dataset.summary()
Overview:
╒===============================================╕
│ Configuration file: │ /mnt/data/datasets.json │
│ Tile size (px): │ 299 │
│ Tile size (um): │ 10x │
│ Slides: │ 941 │
│ Patients: │ 941 │
│ Slides with ROIs: │ 941 │
│ Patients with ROIs: │ 941 │
╘===============================================╛
Filters:
╒====================╕
│ Filters: │ {} │
├--------------------┤
│ Filter Blank: │ [] │
├--------------------┤
│ Min Tiles: │ 0 │
╘====================╛
Sources:
TCGA_LUNG
╒==============================================╕
│ slides │ /mnt/raid/SLIDES/TCGA_LUNG │
│ roi │ /mnt/raid/SLIDES/TCGA_LUNG │
│ tiles │ /mnt/rocket/tiles/TCGA_LUNG │
│ tfrecords │ /mnt/rocket/tfrecords/TCGA_LUNG/ │
│ label │ 299px_10x │
╘==============================================╛
Number of tiles in TFRecords: 18354
Annotation columns:
Index(['patient', 'subtype', 'site', 'slide'],
dtype='object')
Manifest¶
Dataset.manifest() provides a dictionary mapping tfrecords to the total number of image tiles and the number of tiles after clipping or mini-batch balancing. For example, after clipping:
dataset = dataset.clip(500)
the manifest may look something like:
{
"/path/tfrecord1.tfrecords":
{
"total": 1526,
"clipped": 500
},
"/path/tfrecord2.tfrecords":
{
"total": 455,
"clipped": 455
}
}
Inspecting a dataset’s manifest may be useful to better understand the effects of dataset manipulations.
Training/Validation Splitting¶
An important step when planning an experiment is to determine your validation and testing data. In total, deep learning experiments should have three groups of data:
Training - data used for learning during training
Validation - data used for validating training parameters and early stopping (if applicable)
Evaluation - held-out data used for final testing once all training and parameter tuning has completed. Preferably an external cohort.
Slideflow includes tools for flexible training, validation, and evaluation data planning as discussed in the next sections.
Creating a split¶
Datasets can be split into training and validation or test datasets with Dataset.split(). The result of this function is two datasets - the first training, the second validation - each a separate instance of Dataset.
Slideflow provides several options for preparing a validation plan, including:
strategy:
'bootstrap','k-fold','k-fold-manual','k-fold-preserved-site','fixed', and'none'fraction: (float between 0-1) [not used for k-fold validation]
k_fold: int
The default validation strategy is three-fold cross-validation (strategy='k-fold' and k=3).
# Split a dataset into training and validation
# using 5-fold cross-validation, with this being
# the first cross-fold.
train_dataset, test_dataset = dataset.split(
model_type='categorical', # Categorical labels
labels='subtype', # Label to balance between datasets
k_fold=5, # Total number of crossfolds
k_fold_iter=1, # Cross-fold iteration
splits='splits.json' # Where to save/load crossfold splits
)
You can also use Dataset.kfold_split() to iterate through cross-fold splits:
# Split a dataset into training and validation
# using 5-fold cross-validation
for train, test in dataset.kfold_split(k=5, labels='subtype'):
...
Validation strategies¶
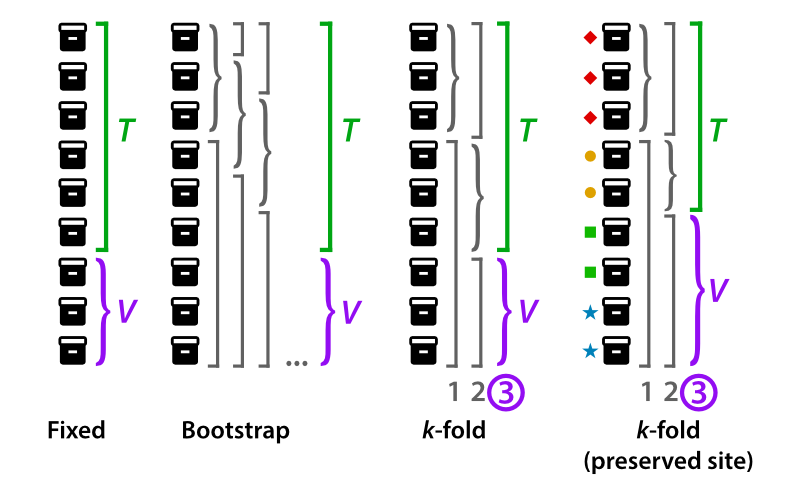
The strategy option determines how the validation data is selected.
If fixed, a certain percentage of your training data is set aside for testing (determined by fraction).
If bootstrap, validation data will be selected at random (percentage determined by fraction), and all training iterations will be repeated a number of times equal to k_fold. When used during training, the reported model training metrics will be an average of all bootstrap iterations.
If k-fold, training data will be automatically separated into k number of groups (where k is equal to k_fold), and all training iterations will be repeated k number of times using k-fold cross validation. The saved and reported model training metrics will be an average of all k-fold iterations.
Datasets can be separated into manually-curated k-folds using the k-fold-manual strategy. Assign each slide to a k-fold cohort in the annotations file, and designate the appropriate column header with k_fold_header
The k-fold-preserved-site strategy is a cross-validation strategy that ensures site is preserved across the training/validation sets, in order to reduce bias from batch effect as described by Howard, et al. This strategy is recommended when using data from The Cancer Genome Atlas (TCGA).
Note
Preserved-site cross-validation requires either CPLEX or Pyomo/Bonmin. The original implementation of the preserved-site cross-validation algorithm described by Howard et al can be found on GitHub.
If none, no validation testing will be performed.
Re-using splits¶
For all validation strategies, training/validation splits can be logged to a JSON file automatically if a splits configuration file is provided to the argument splits. When provided, Dataset.split() will prioritize using previously-generated training/validation splits rather than generating a new split. This aids with experiment reproducibility and hyperparameter tuning. If training/validation splits are being prepared by a Project-level function, splits will be automatically logged to a splits.json file in the project root directory.
Creating Dataloaders¶
Finally, Datasets can also return either a tf.data.Datasets or torch.utils.data.Dataloader object to quickly and easily create a deep learning dataset ready to be used as model input, with the Dataset.tensorflow() and Dataset.torch() methods, respectively. See Dataloaders: Sampling and Augmentation for more detailed information and examples.
Datasets have many other utility functions for working with and processing data. Read more in the Dataset API documentation.