Multiple-Instance Learning (MIL)¶
In addition to standard tile-based neural networks, Slideflow also supports training multiple-instance learning (MIL) models. Several architectures are available, including attention-based MIL ("Attention_MIL"), CLAM ("CLAM_SB", "CLAM_MB", "MIL_fc", "MIL_fc_mc"), TransMIL ("TransMIL"), and HistoBistro Transformer ("bistro.transformer"). Custom architectures can also be trained. MIL training requires PyTorch.
Skip to Tutorial 8: Multiple-Instance Learning for a complete example of MIL training.
See slideflow.mil for more information on the MIL API.
Generating Features¶
The first step in MIL model development is generating features from image tiles, as discussed in the Generating Features section. Features from whole-slide images are exported as “bags” of features, where each bag contains a set of features from a single slide. Each bag is a PyTorch tensor saved in *.pt format. Bags are saved in a directory, and the directory path is passed to the MIL model during training and evaluation.
Training¶
Model Configuration¶
To train an MIL model using exported features, first prepare an MIL configuration using slideflow.mil.mil_config().
The first argument to this function is the model architecture (which can be a name or a custom torch.nn.Module model), and the remaining arguments are used to configure the training process, such as learning rate and number of epochs. Training is executed using FastAI with 1cycle learning rate scheduling.
import slideflow as sf
from slideflow.mil import mil_config
config = mil_config('attention_mil', lr=1e-3)
Available models out-of-the-box include attention-based MIL ("Attention_MIL"), transformer MIL ("TransMIL"), and HistoBistro Transformer ("bistro.transformer"). CLAM ("CLAM_SB", "CLAM_MB", "MIL_fc", "MIL_fc_mc") models are available through slideflow-gpl:
pip install slideflow-gpl
Custom MIL models can also be trained with this API, as discussed below.
Classification & Regression¶
MIL models can be trained for both classification and regression tasks. The type of outcome is determined through the loss function, which defaults to "cross_entropy". To train a model for regression, set the loss function to one of the following regression losses, and ensure that your outcome labels are continuous. You can also train to multiple outcomes by passing a list of outcome names.
“mse” (
nn.CrossEntropyLoss): Mean squared error.“mae” (
nn.L1Loss): Mean absolute error.“huber” (
nn.SmoothL1Loss): Huber loss.
# Prepare a regression-compatible MIL configuration
config = mil_config('attention_mil', lr=1e-3, loss='mse')
# Train the model
project.train_mil(
config=config,
...,
outcomes=['age', 'grade']
)
Training an MIL Model¶
Next, prepare a training and validation dataset and use slideflow.Project.train_mil() to start training. For example, to train a model using three-fold cross-validation to the outcome “HPV_status”:
...
# Prepare a project and dataset
P = sf.Project(...)
full_dataset = dataset = P.dataset(tile_px=299, tile_um=302)
# Split the dataset using three-fold, site-preserved cross-validation
splits = full_dataset.kfold_split(
k=3,
labels='HPV_status',
preserved_site=True
)
# Train on each cross-fold
for train, val in splits:
P.train_mil(
config=config,
outcomes='HPV_status',
train_dataset=train,
val_dataset=val,
bags='/path/to/bag_directory'
)
Model training statistics, including validation performance (AUROC, AP) and predictions on the validation dataset, will be saved in an mil subfolder within the main project directory.
If you are training an attention-based MIL model (attention_mil, clam_sb, clam_mb), heatmaps of attention can be generated for each slide in the validation dataset by using the argument attention_heatmaps=True. You can customize these heatmaps with interpolation and cmap arguments to control the heatmap interpolation and colormap, respectively.
# Generate attention heatmaps,
# using the 'magma' colormap and no interpolation.
P.train_mil(
attention_heatmaps=True,
cmap='magma',
interpolation=None
)
Hyperparameters, model configuration, and feature extractor information is logged to mil_params.json in the model directory. This file also contains information about the input and output shapes of the MIL network and outcome labels. An example file is shown below.
{
"trainer": "fastai",
"params": {
},
"outcomes": "histology",
"outcome_labels": {
"0": "Adenocarcinoma",
"1": "Squamous"
},
"bags": "/mnt/data/projects/example_project/bags/simclr-263510/",
"input_shape": 1024,
"output_shape": 2,
"bags_encoder": {
"extractor": {
"class": "slideflow.model.extractors.simclr.SimCLR_Features",
"kwargs": {
"center_crop": false,
"ckpt": "/mnt/data/projects/example_project/simclr/00001-EXAMPLE/ckpt-263510.ckpt"
}
},
"normalizer": null,
"num_features": 1024,
"tile_px": 299,
"tile_um": 302
}
}
Multi-Magnification MIL¶
Slideflow 2.2 introduced a multi-magnification, multi-modal MIL model, MultiModal_Attention_MIL ("mm_attention_mil"). This late-fusion multimodal model is based on standard attention-based MIL, but accepts multiple input modalities (e.g., multiple magnifications) simultaneously. Each input modality is processed by a separate encoder network and a separate attention module. The attention-weighted features from each modality are then concatenated and passed to a fully-connected layer.
Multimodal models are trained using the same API as standard MIL models. Modalities are specified using the bags argument to slideflow.Project.train_mil(), where the number of modes is determined by the number of bag directories provided. Within each bag directory, bags should be generated using the same feature extractor and at the same magnification, but feature extractors and magnifications can vary between bag directories.
For example, to train a multimodal model using two magnifications, you would pass two bag paths to the model. In this case, the /path/to/bags_10x directory contains bags generated from a 10x feature extractor, and the /path/to/bags_40x directory contains bags generated from a 40x feature extractor.
# Configure a multimodal MIL model.
config = mil_config('mm_attention_mil', lr=1e-4)
# Set the bags paths for each modality.
bags_10x = '/path/to/bags_10x'
bags_40x = '/path/to/bags_40x'
P.train_mil(
config=config,
outcomes='HPV_status',
train_dataset=train,
val_dataset=val,
bags=[bags_10x, bags_40x]
)
You can use any number of modalities, and the feature extractors for each modality can be different. For example, you could train a multimodal model using features from a custom SimCLR model at 5x and features from a pretrained CTransPath model at 20x.
The feature extractors used for each modality, as specified in the bags_config.json files in the bag directories, will be logged in the final mil_params.json file. Multimodal MIL models can be interactively viewed in Slideflow Studio, allowing you to visualize the attention weights for each modality separately.
Custom Architectures¶
Training custom MIL models is straightforward with Slideflow, particularly if your model can adhere to a few simple guidelines:
Initialized with
(num_feats, num_outputs)(e.g.,Attention_MIL(768, 2))Input is feature bags with shape
(batch, num_tiles, num_feats). If the model needs a “lens” input, then the model attributeuse_lensshould be True.Has a
relocate()function that moves the model to detected device/GPU- Ability to get attention through one of two methods:
forward()function includes an optionalreturn_attentionargument, which if True returns attention scores after model outputHas a
calculate_attention()function that returns attention scores
If the above applies to your model, you can train it simply by passing it as the first argument to slideflow.mil.mil_config().
import slideflow as sf
from slideflow.mil import mil_config
from my_module import CustomMIL
config = mil_config(CustomMIL, lr=1e-3)
For larger projects, or if you are designing a plugin/extension for Slideflow, custom models can be registered to facilitate easy creation. If your model adheres to the above guidelines, you can register it for use with the following:
from slideflow.mil import register_model
@register_model
def my_model():
return MyModelClass
You can then use your model when creating an MIL configuration:
config = sf.mil.mil_config('my_model', ...)
If the above guidelines do not apply to your model, or if you want to customize model logic or functionality, you can supply a custom MIL configuration class that will supervise model building and dataset preparation. Your custom configuration class should inherit slideflow.mil.MILModelConfig, and methods in this class can be overloaded to provide additional functionality. For example, to create an MIL configuration that uses a custom loss and custom metrics:
from slideflow.mil import MILModelConfig
class MyModelConfig(MILModelConfig):
@property
def loss_fn(self):
return my_custom_loss
def get_metrics(self):
return [my_metric1, my_metric2]
When registering your model, you should specify that it should use your custom configuration:
@register_model(config=MyModelConfig)
def my_model():
return MyModelClass
For an example of how to utilize model registration and configuration customization, see our CLAM implementation available through slideflow-gpl.
Evaluation¶
To evaluate a saved MIL model on an external dataset, first extract features from a dataset, then use slideflow.Project.evaluate_mil(), which displays evaluation metrics and returns predictions as a DataFrame.
import slideflow as sf
# Prepare a project and dataset
P = sf.Project(...)
dataset = P.dataset(tile_px=299, tile_um=302)
# Generate features using CTransPath
ctranspath = sf.build_feature_extractor('ctranspath', resize=True)
features = sf.DatasetFeatures(ctranspath, dataset=dataset)
features.to_torch('/path/to/bag_directory')
# Evaluate a saved MIL model
df = P.evaluate_mil(
'/path/to/saved_model'
outcomes='HPV_status',
dataset=dataset,
bags='/path/to/bag_directory',
)
As with training, attention heatmaps can be generated for attention-based MIL models with the argument attention_heatmaps=True, and these can be customized using cmap and interpolation arguments.
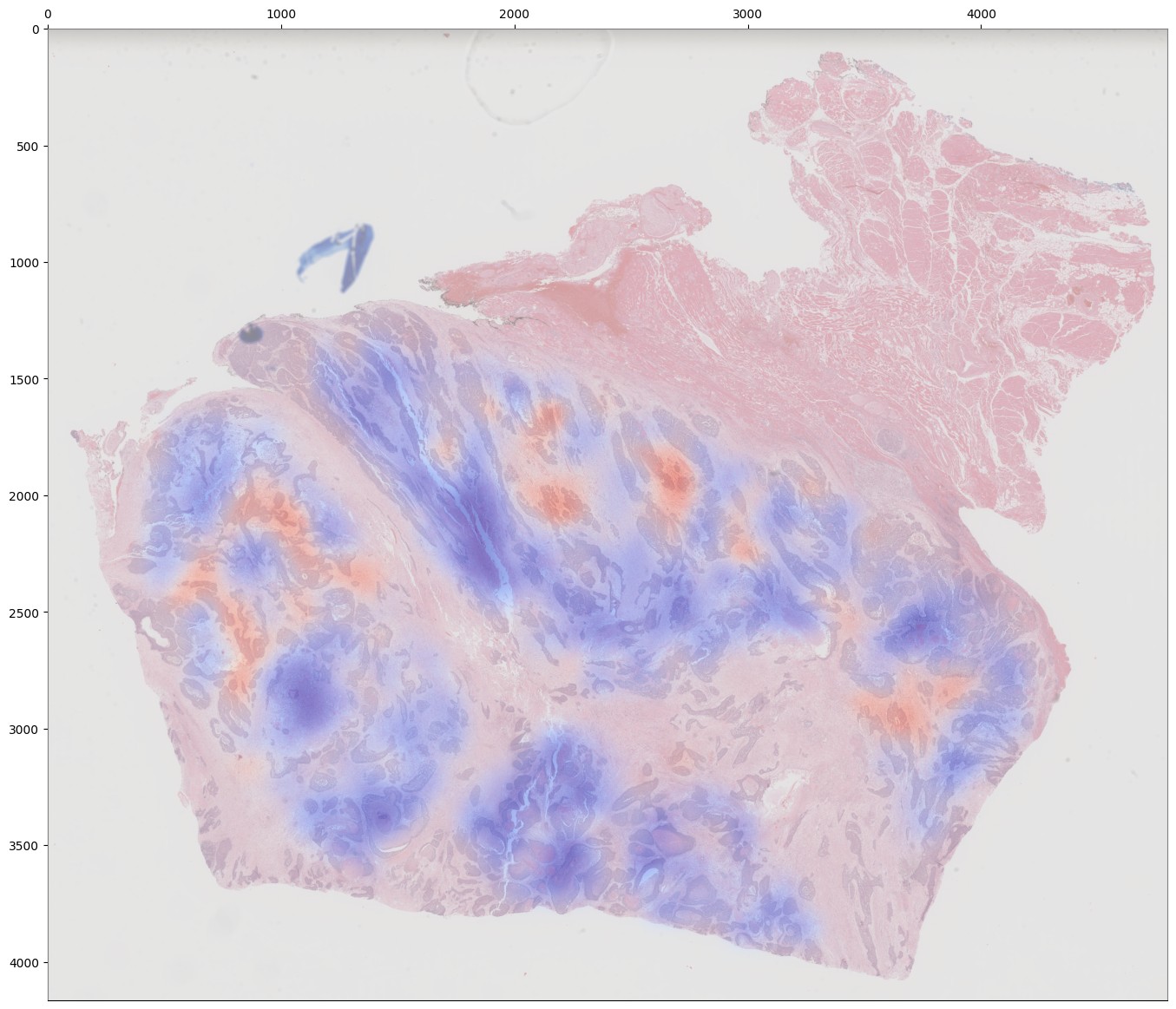
Generating Predictions¶
In addition to generating slide-level predictions during training and evaluation, you can also generate tile-level predictions and attention scores for a dataset using slideflow.mil.get_mil_tile_predictions(). This function returns a DataFrame containing tile-level predictions and attention.
>>> from slideflow.mil import get_mil_tile_predictions
>>> df = get_mil_tile_predictions(model, dataset, bags)
>>> df
slide loc_x loc_y ... y_pred3 y_pred4 y_pred5
0 TCGA-4V-A9QI-01Z-0... 2210 7349 ... 0.181155 0.468446 0.070175
1 TCGA-4V-A9QI-01Z-0... 5795 1971 ... 0.243721 0.131991 0.009169
2 TCGA-4V-A9QI-01Z-0... 6273 5437 ... 0.096196 0.583367 0.090258
3 TCGA-4V-A9QI-01Z-0... 2330 3047 ... 0.056426 0.264386 0.300199
4 TCGA-4V-A9QI-01Z-0... 3644 3525 ... 0.134535 0.534353 0.013619
... ... ... ... ... ... ... ...
391809 TCGA-4X-A9FA-01Z-0... 6034 3352 ... 0.004119 0.003636 0.005673
391810 TCGA-4X-A9FA-01Z-0... 6643 1401 ... 0.012790 0.010269 0.011726
391811 TCGA-4X-A9FA-01Z-0... 5546 2011 ... 0.009777 0.013556 0.025255
391812 TCGA-4X-A9FA-01Z-0... 6277 2864 ... 0.026638 0.018499 0.031061
391813 TCGA-4X-A9FA-01Z-0... 4083 4205 ... 0.009875 0.009582 0.022125
[391814 rows x 15 columns]
Single-Slide Inference¶
Predictions can also be generated for individual slides, without requiring the user to manually generate feature bags. Use slideflow.model.predict_slide() to generate predictions for a single slide. The first argument is th path to the saved MIL model (a directory containing mil_params.json), and the second argument can either be a path to a slide or a loaded sf.WSI object.
from slideflow.mil import predict_slide
from slideflow.slide import qc
# Load a slide and apply Otsu thresholding
slide = '/path/to/slide.svs'
wsi = sf.WSI(slide, tile_px=299, tile_um=302)
wsi.qc(qc.Otsu())
# Calculate predictions and attention heatmap
model = '/path/to/mil_model'
y_pred, y_att = predict_slide(model, wsi)
The function will return a tuple of predictions and attention heatmaps. If the model is not attention-based, the attention heatmap will be None. To calculate attention for a model, set attention=True:
y_pred, y_att = predict_slide(model, slide, attention=True)
The returned attention values will be a masked numpy.ndarray with the same shape as the slide tile extraction grid. Unused tiles will have masked attention values.
Visualizing Predictions¶
Heatmaps of attention and tile-level predictions can be interactively visualized in Slideflow Studio by enabling the Multiple-Instance Learning extension (new in Slideflow 2.1.0). This extension is discussed in more detail in the Extensions section.